Grouping data¶
A single pandas DataFrame can often be decomposed into smaller subsets of data that need to be visualized. fivecentplots contains multiple grouping levels to expose unique subsets of data:
legend: color lines and markers according to unique values in a DataFrame column
groups:
for xy plots: separates unique subsets so plot lines are not continuously looped back to the origin (useful for replicates of similar data)
for boxplots: groups boxes by unique values in one or more DataFrame columns
row | column: makes a grid of subplots for each unique value of the DataFrame column names specified for these keyword arguments within a single figure
wrap:
Option 1: similar to row and column grouping, wrap makes a grid of subplots for each unique value of the DataFrame column names specified for these keyword arguments within a single figure
Option 2: wrap by
xoryto create a uniqe subplot for each column name listed
figure: makes a unique figure for each unique value of a DataFrame column
It is also possible to filter data inline sets via the keyword filter.
Setup¶
Imports¶
In [2]:
%load_ext autoreload
%autoreload 2
%matplotlib inline
import fivecentplots as fcp
import pandas as pd
import numpy as np
import os, sys, pdb
osjoin = os.path.join
st = pdb.set_trace
Sample data¶
In [3]:
df1 = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data.csv'))
df2 = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data_box.csv'))
legend¶
The legend keyword can be a single DataFrame column name or a list
of column names. The data set will then be grouped according to each
unique value of the legend column and a separate plot for each value
will be added to the figure. A different color and marker type will be
used to display each value.
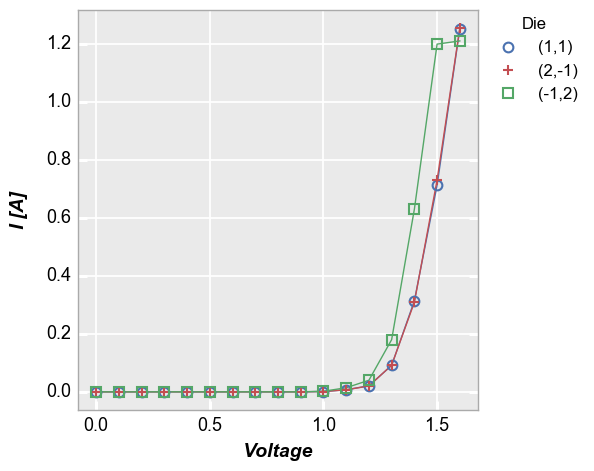
Single legend column¶
In our sample data set, we have repeats of the same current vs. voltage
measurement at three different “Die” locations. By setting the
legend keyword equal to the name of the DataFrame column containing
the “Die” value, we can distinctly visualize the measurement for each
site.
In [8]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Die',
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25')
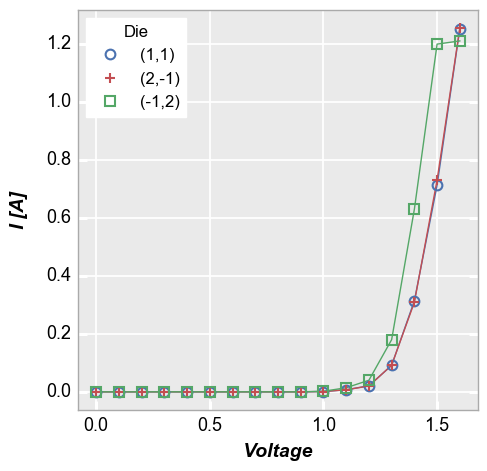
Multiple legend columns¶
fivecentplots also supports legending by multiple DataFrame columns.
When a list of column names is passed to the legend keyword, a dummy
column is created in the DataFrame that concatenates the values from
each column listed in legend. This new column is used for legending.
In [4]:
fcp.plot(df1, x='Voltage', y='I [A]', legend=['Die', 'Substrate'], \
filter='Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25')
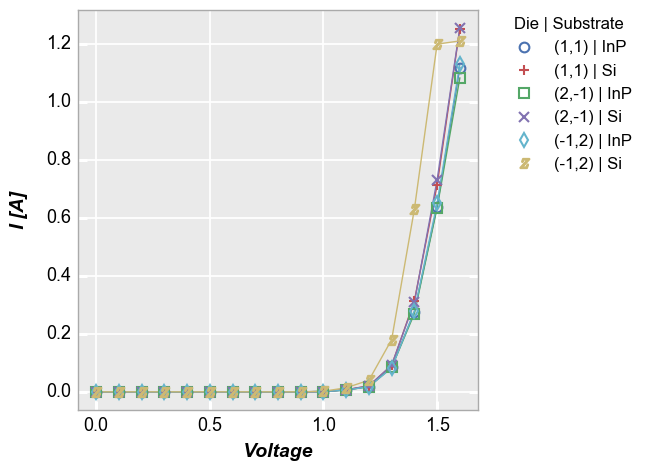
Multiple x & y values¶
When plotting more than one DataFrame column on the y axis without a
specific grouping column, a legend is also enabled to improve
visualization of the data. The legend can be disabled by setting
legend=False.
In [5]:
fcp.plot(df1, x='Voltage', y=['I [A]', 'Voltage'], lines=False,
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2')
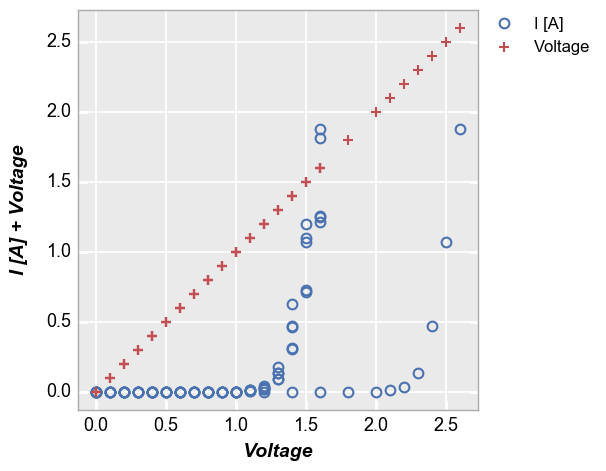
Secondary x|y plots¶
Three options are available for legending are available for plots with a secondary axis: (1) no legend; (2) legend based on the values of the primary/secondary axes; or (3) legend based on another column.
No legend¶
In [6]:
fcp.plot(df1, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True,
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"')
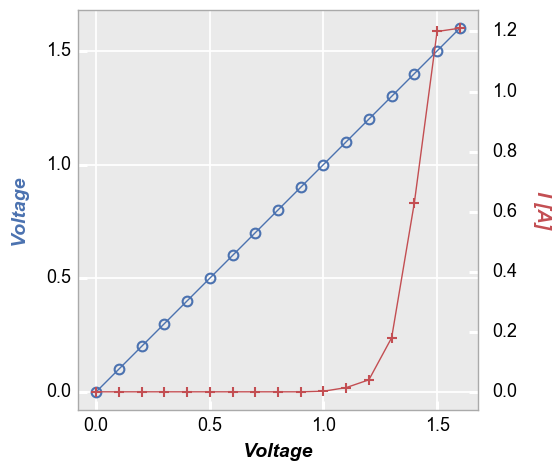
Axis legend¶
In [7]:
fcp.plot(df1, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, legend=True,
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"')
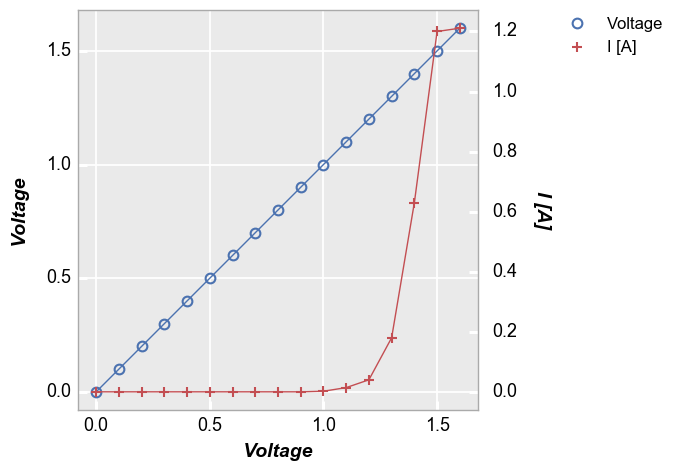
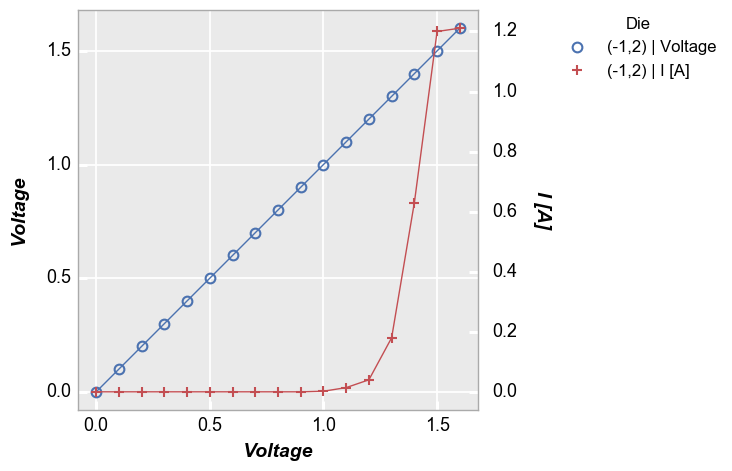
Legend by another column¶
In [8]:
fcp.plot(df1, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, legend='Die',
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"')
Location¶
By default, fivecentplots places the legend in the upper right corner of
a figure, outside of the plot axes as this is the only way to be certain
the legend will never obscure the actual data. However, the legend can
be placed inside the axes area by specifying the keyword
legend_location with one of the following values (which align
roughly matplotlib syntax):
- ‘outside’ or 0
- ‘upper right’ or 1
- ‘upper left’ or 2
- ‘lower left’ or 3
- ‘lower right’ or 4
- ‘right’ or 5
- ‘center left’ or 6
- ‘center right’ or 7
- ‘lower center’ or 8
- ‘upper center’ or 9
- ‘center’ or 10
Note this feature is not currently supported with the bokeh plotting
engine
In [4]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Die',
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25', legend_location=2)
Relocating the legend is straightforward when dealing with a single plot
axes, but the ideal situation is a bit murky when dealing with grid
plots with legends (see below). By default, the legend will be placed in
each subplot window. However, if the keyword nleg is included and
set equal to 1, the legend will appear in the new location but only
within the upper right subplot.
groups¶
The groups keyword requires a column name or list of column names
from the DataFrame. These columns are used to identify subsets in the
data and make sure they are properly visualize in the plot. Unlike
legend, group values are not added to a legend on the plot.
xy plots¶
Some data sets contain multiple sets of similar data. Consider the following example where we plot all the data together and connect the points with lines. Notice how the line loops from the end of the data back to the begining for each “group” of the data. This occurs because we have provided no information about how to segment the data set.
In [9]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Temperature [C]', \
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2')
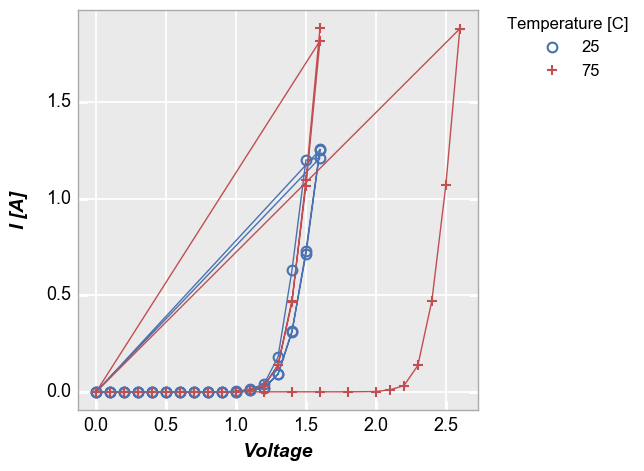
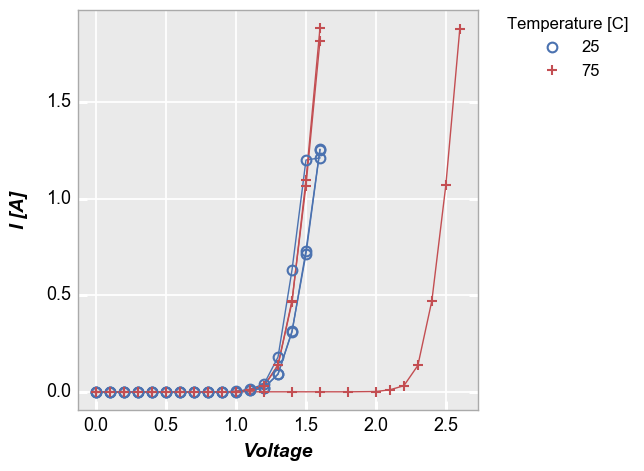
To handle cases like this, we can add the keyword groups and specify
another DataFrame column name that indicates how the data are grouped
(in this case by “Die”). Now we get distinct lines for each instance of
the measurement data. The groups keyword can still be combined with
a legend from a different DataFrame column.
In [10]:
fcp.plot(df1, x='Voltage', y='I [A]', groups='Die', legend='Temperature [C]', \
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2')
groups also supports multiple column names. Here is the above plot
without legending by “Temperature [C]”:
In [11]:
fcp.plot(df1, x='Voltage', y='I [A]', groups=['Die', 'Temperature [C]'],
filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2')
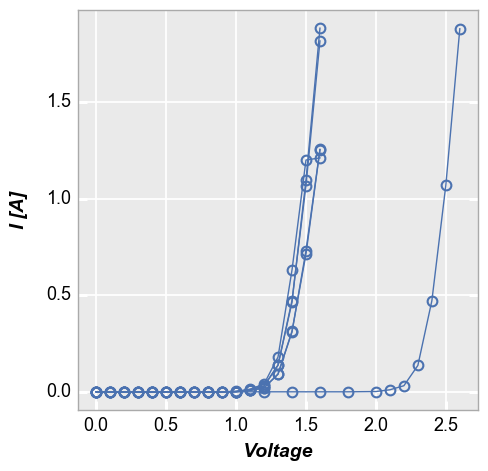
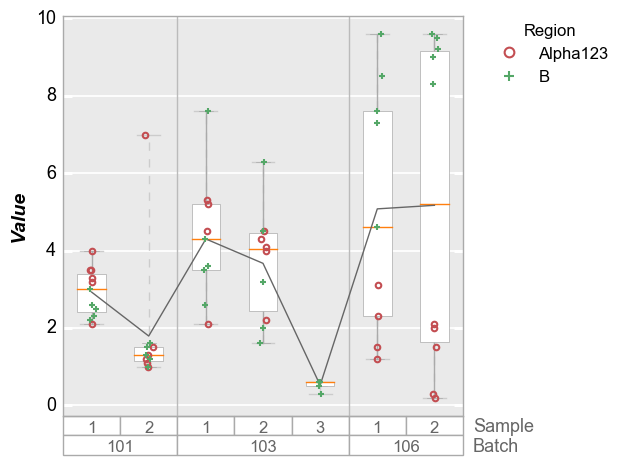
boxplots¶
Like x-y plots, the groups keyword is used to break the data set
into subsets. However, for boxplots the group column names and values
are actually displayed along the x-axis:
In [12]:
df_box = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data_box.csv'))
fcp.boxplot(df=df_box, y='Value', groups=['Batch', 'Sample'], legend='Region')
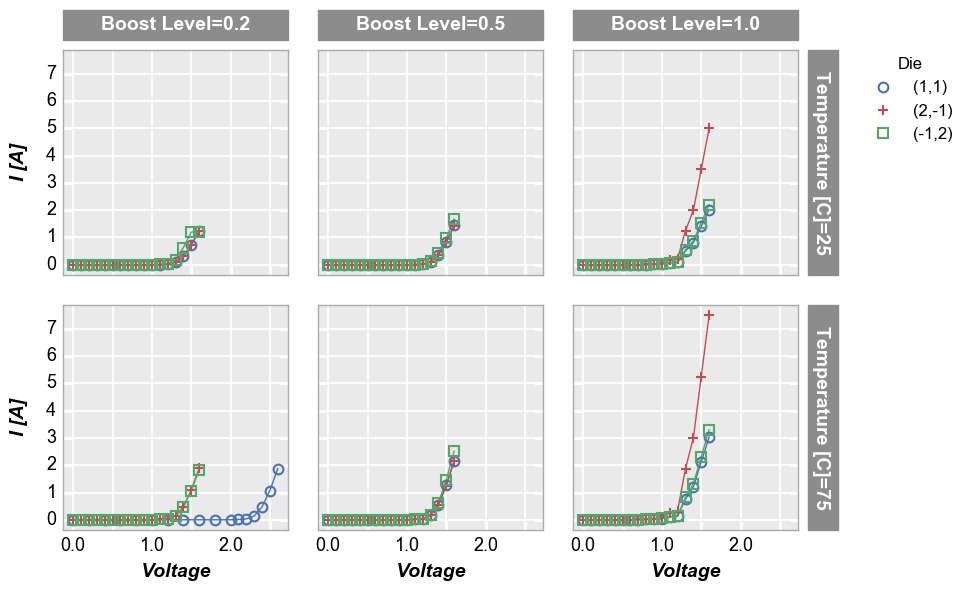
row | col subplots¶
By unique values¶
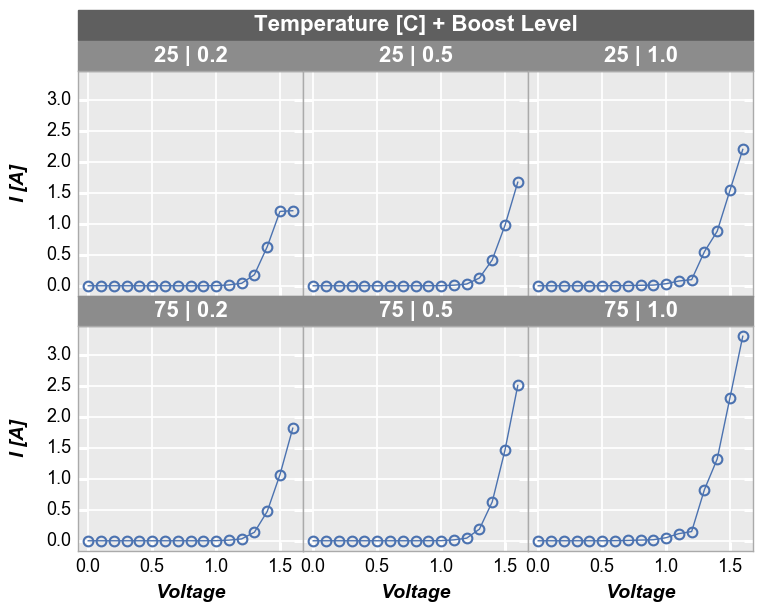
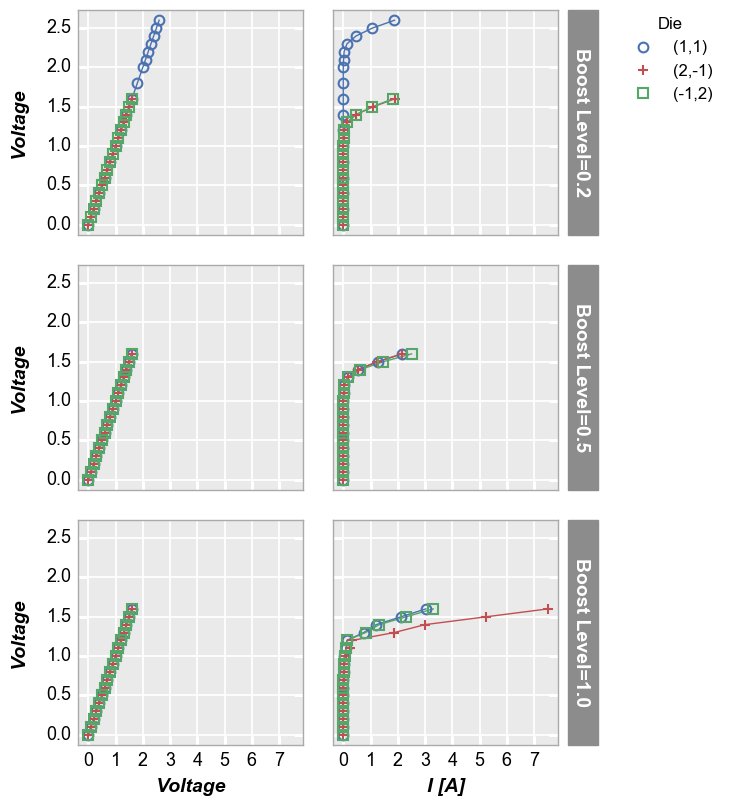
To see a larger subset of the main DataFrame, we can make a grid of subplots based on the unique values within DataFrame columns other than the primary x and y columns we are using. In this case, we remove the “Temperature [C]” and “Boost Level” columns from the filter keyword and add them to the row and column commands, respectively. This gives a grid of plots where each plot represents the intersection of a unique Boost Level and Temperature.
In [13]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Die', col='Boost Level', row='Temperature [C]', \
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450', label_rc_font_size=14)
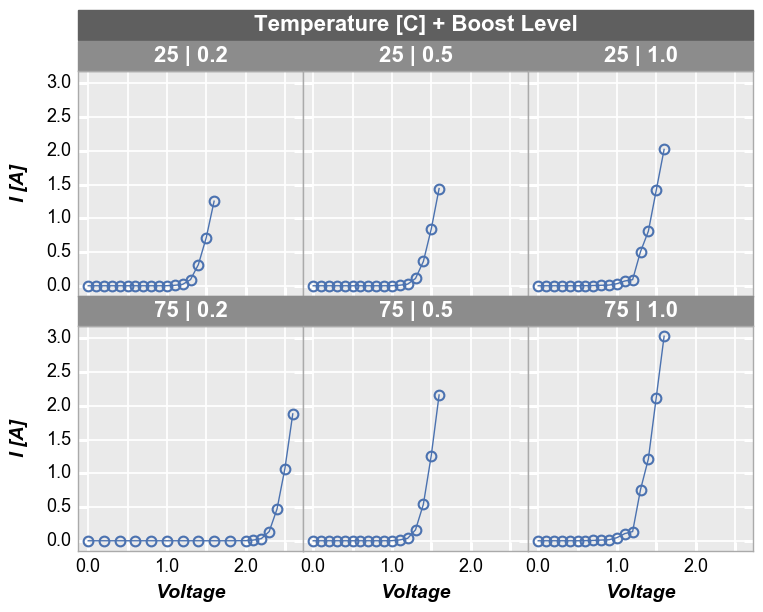
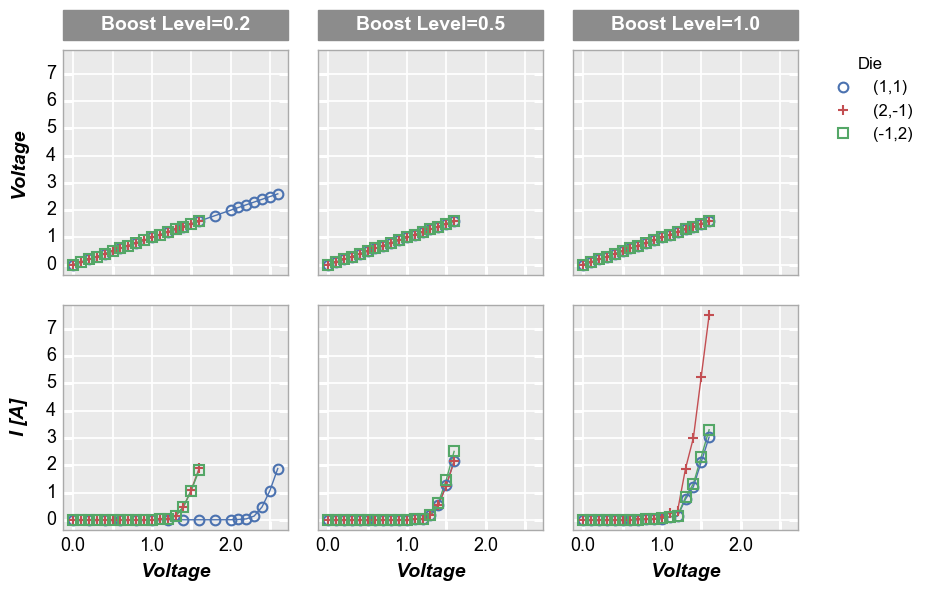
By x or y¶
Alternatively, we can use row vs column grids to compare different x
or y values. For example, we can plot two y-columns (one per row) by
adding the keyword "y" to the row parameter. Each column in this
plot shares the same x-axis variable.
In [14]:
fcp.plot(df1, x='Voltage', y=['Voltage', 'I [A]'], legend='Die', col='Boost Level', row='y',
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450 & Temperature [C]==75',
label_rc_font_size=14)
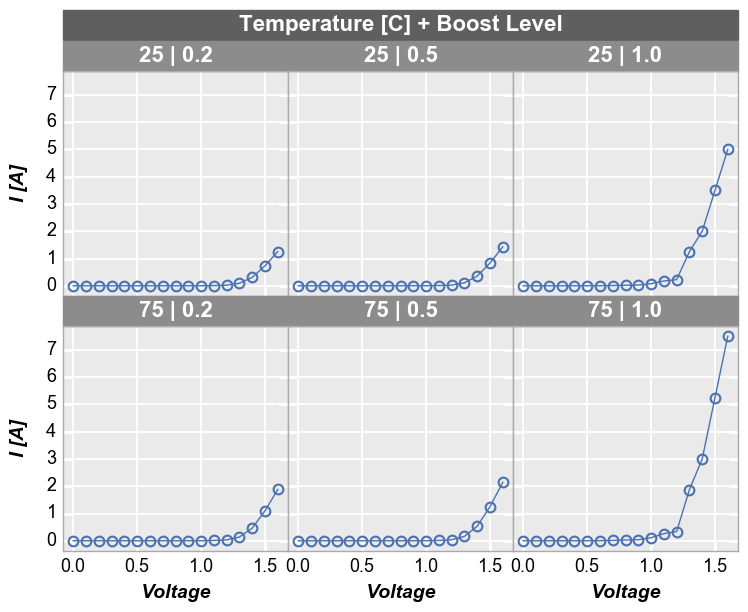
We can make a similar plot with common row values and different
x-axes by setting col='x':
In [15]:
fcp.plot(df1, x=['Voltage', 'I [A]'], y='Voltage', legend='Die', row='Boost Level', col='x',
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450 & Temperature [C]==75', label_rc_font_size=14)
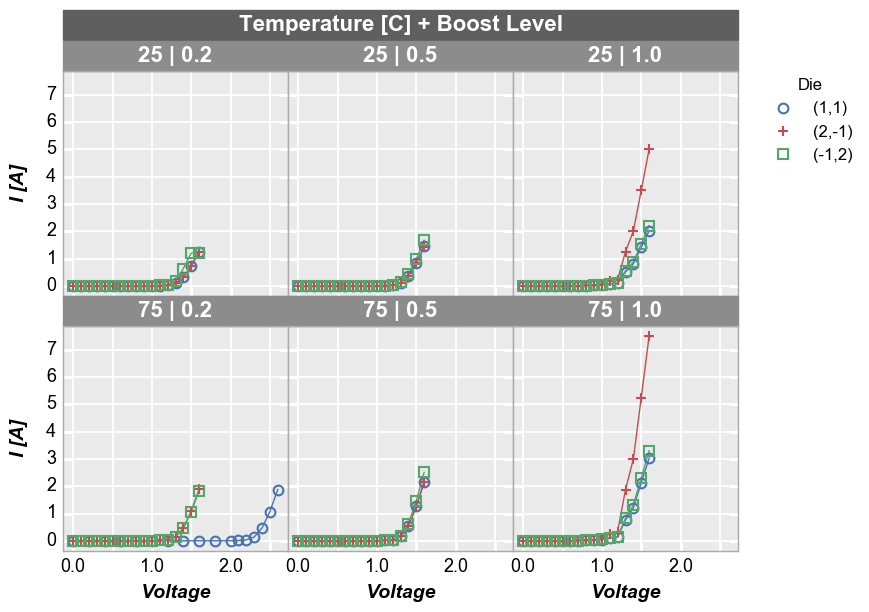
wrap subplots¶
By unique values¶
We can create a plot similar to the row/column plot above using the
wrap keyword. The key differences are that spacing and ticks between
subplots is removed by default (can be changed via keywords), axis
sharing is forced (cannot override), and the row/column value labels are
condensed to a single label above each subplot. To do this, remove the
row and column keywords from the previous function call and pass
the “Temperature [C]” and “Boost Level” column names as a list to the
wrap keyword.
In [16]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Die', wrap=['Temperature [C]', 'Boost Level'], \
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450')
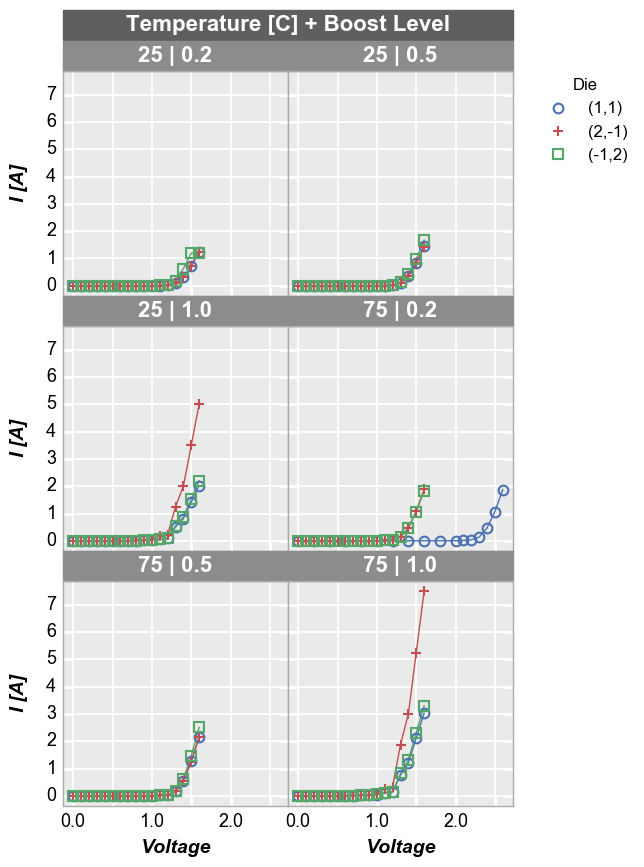
By default, wrap plots will be arranged in a square grid. If the number
of subplots does not match a perfectly square grid, you will have an
incomplete row of subplots in the grid. All tick and axes labels are
handled appropriately for this case. If you want to override the default
grid size, specify the keyword ncol which sets the number of
columns. The number of rows will be automatically determined from the
number of subplots.
In [17]:
fcp.plot(df1, x='Voltage', y='I [A]', legend='Die', wrap=['Temperature [C]', 'Boost Level'], \
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450', ncol=2)
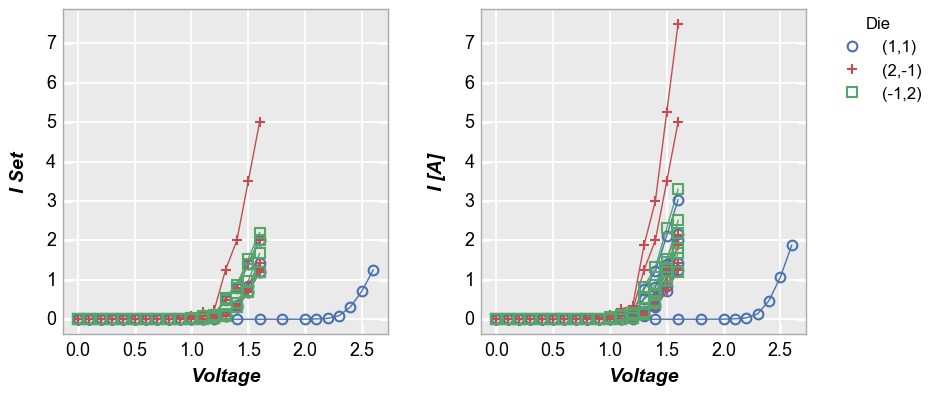
By x and y¶
We can also “wrap” by column names instead of unique column values. In
this case we list the columns to plot in the x or y keywords as
usual but we add “x” or “y” to the wrap keyword. This will create a
unique subplot in a grid for each wrap value. Unlike the case of
wrapping column values, this plot will not have wrap labels and axis
sharing can be overriden. As before, used ncol to override the
default gridding. Unlike a row vs column plot by x and y, a wrap plot by
x and y can be displayed without the extra grouping factor.
In [18]:
fcp.plot(df1, x='Voltage', y=['I Set', 'I [A]'], legend='Die', wrap='y',
groups=['Boost Level', 'Temperature [C]'], ax_size=[325, 325],
filter='Substrate=="Si" & Target Wavelength==450')
Or an alternatively styled version of this plot with tick and label sharing off:
In [19]:
fcp.plot(df1, x='Voltage', y=['I Set', 'I [A]'], legend='Die', wrap='y',
groups=['Boost Level', 'Temperature [C]'], ax_size=[525, 170],
filter='Substrate=="Si" & Target Wavelength==450', ncol=1, ws_row=0,
separate_labels=False, separate_ticks=False)
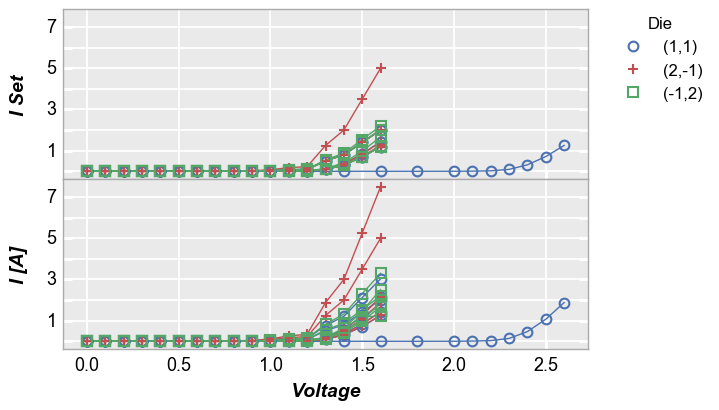
figure plots¶
To add another dimension of grouping, fivecentplots supports grouping by
figure. In this case, a separate figure (i.e., a separate png) is
created for each unique value in the DataFrame column(s) listed in the
fig_groups keyword. Here we will plot a single figure for each value
of the “Die” column.
In [11]:
fcp.plot(df1, x='Voltage', y='I [A]', fig_groups='Die', wrap=['Temperature [C]', 'Boost Level'], \
ax_size=[225, 225], filter='Substrate=="Si" & Target Wavelength==450', show_filename=True,
filename=r'C:\GitHub\fivecentplots\fivecentplots\tests\test_images\grouping.py\figure.png')