Data ranges¶
This section provides examples of how to set data ranges for a plot via keyword arguments. Options include:
automatically calculating limits based on a percentage of the total data range
explicitly setting limits for a given axis
setting limits based on a quantile statistic
sharing or not sharing limits across subplots
Setup¶
Imports¶
In [1]:
%load_ext autoreload
%autoreload 2
%matplotlib inline
import fivecentplots as fcp
import pandas as pd
import numpy as np
import os, sys, pdb
osjoin = os.path.join
st = pdb.set_trace
Read sample data¶
In [2]:
df = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data.csv'))
df_box = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data_box.csv'))
Set theme¶
In [3]:
#fcp.set_theme('gray')
#fcp.set_theme('white')
Other¶
In [4]:
SHOW = False
Default limits¶
When no limits are specified, fivecentplots will attempt to choose reasonable axis limits automatically. This is done by subtracting or adding a percentage of the total data range to the minimum or maximum limit, respectively. Consider the following example:
In [5]:
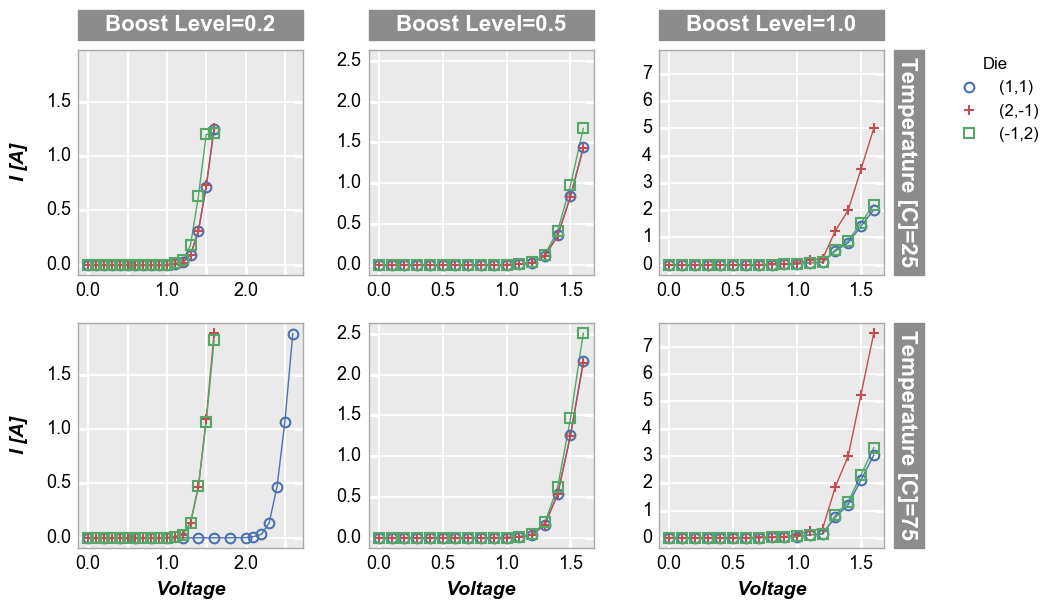
sub = df[(df.Substrate=='Si')&(df['Target Wavelength']==450)&(df['Boost Level']==0.2)&(df['Temperature [C]']==25)]
print('xmin=%s, xmax=%s' % (sub['Voltage'].min(), sub['Voltage'].max()))
print('ymin=%s, ymax=%s' % (sub['I [A]'].min(), sub['I [A]'].max()))
fcp.plot(df=sub, x='Voltage', y='I [A]', legend='Die', show=SHOW)
xmin=0.0, xmax=1.6
ymin=0.0, ymax=1.255
Notice the actual x data range goes from 0 to 1.6 but the x-limits
on the plot go from -0.08 to 1.68 or 5% beyond the x-range. By default,
fivecentplots uses a 5% buffer on non-specified axis limits. For a
log-scaled axis, the data range is calculated as
np.log10(max_val) - np.log10(min_val) to ensure an effective 5%
buffer on the linear-scale of the plot window. This percentage can be
customized by keyword argument or in the theme file by setting
ax_limit_padding to a percentage value divided by 100. Additionally,
the padding limit can be set differently for each axis by including the
axis name and min/max target in the keyword (such as
ax_limit_padding_x_min.
Explicit limits¶
In many cases we want to plot data over a specific data range. This is accomplished by passing set limit values as keywords in the plot command. The following axis can be specified:
x(primary x-axis)x2(secondary x-axis whentwin_y=True)y(primary y-axis)y2(secondary y-axis whentwin_x=True)</li> <li>x(primary z-axis [for heatmaps and contours this is the colorbar axis])</li> </ul> Each axis has aminor amax` value that can be specified.Primary axes only¶
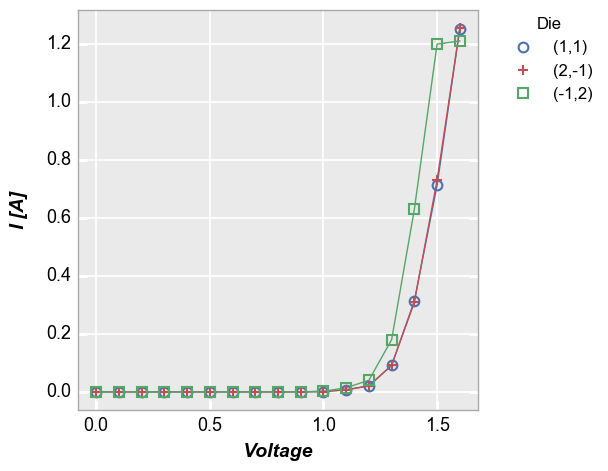
Let’s take the plot from above and zoom in to exclude most of the region where the current begins to grow exponentially. We can do this by only specifying an
xmaxlimit:In [6]:
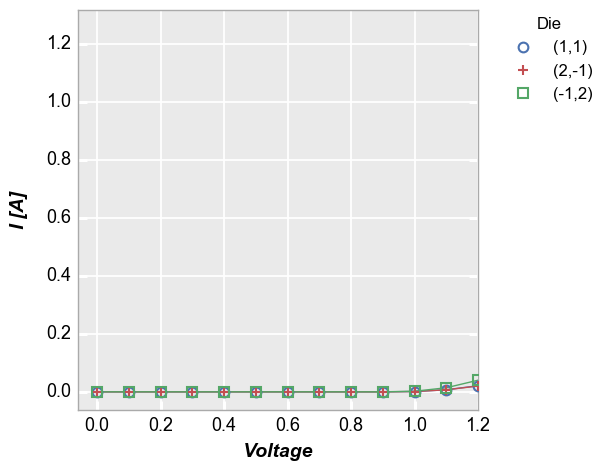
fcp.plot(df=df, x='Voltage', y='I [A]', legend='Die', show=SHOW, filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25', xmax=1.2)
Notice that although we only specified a single limit, the y-axis range has been auto-scaled to more clearly show the data that is included in the x-axis range on interest. This scaling is controlled by the keyword
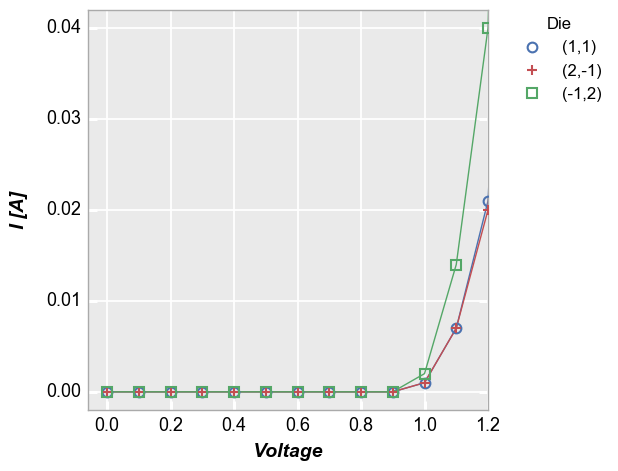
auto_scalewhich is enabled by default. Without auto-scaling the plot would look as follows:In [7]:
fcp.plot(df=df, x='Voltage', y='I [A]', legend='Die', show=SHOW, filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25', xmax=1.2, auto_scale=False)
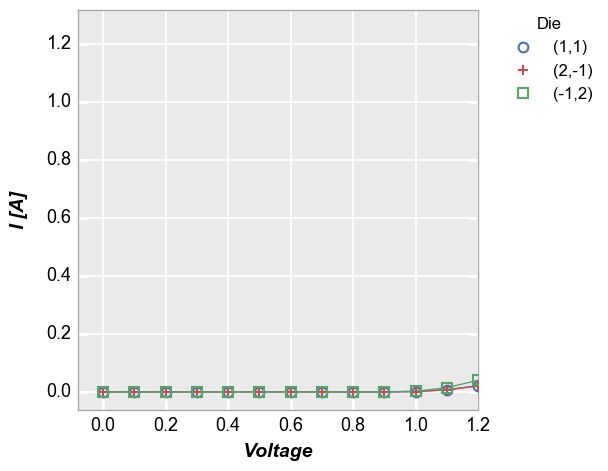
We can accomplish the same thing with
auto_scale=Trueif we specify the y-axis range explictly (note that we are including theax_limit_paddingof 0.05 to match exactly):In [8]:
fcp.plot(df=df, x='Voltage', y='I [A]', legend='Die', show=SHOW, filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25', xmax=1.2, ymin=-0.06275, ymax=1.31775)
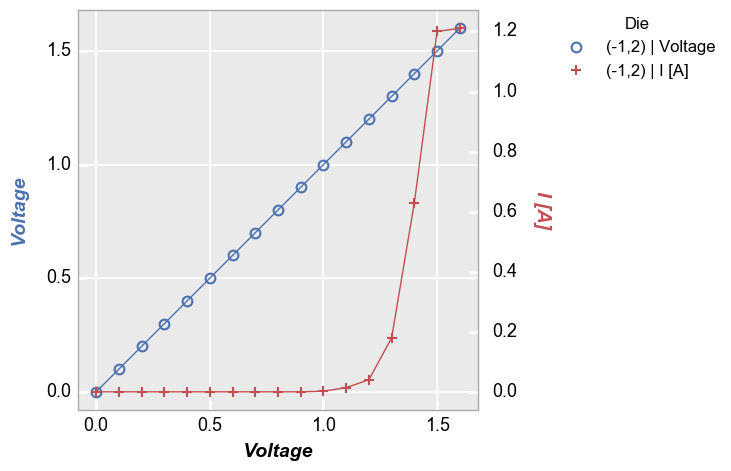
Secondary y-axis¶
Now condsider the case of a secondary y-axis:
In [9]:
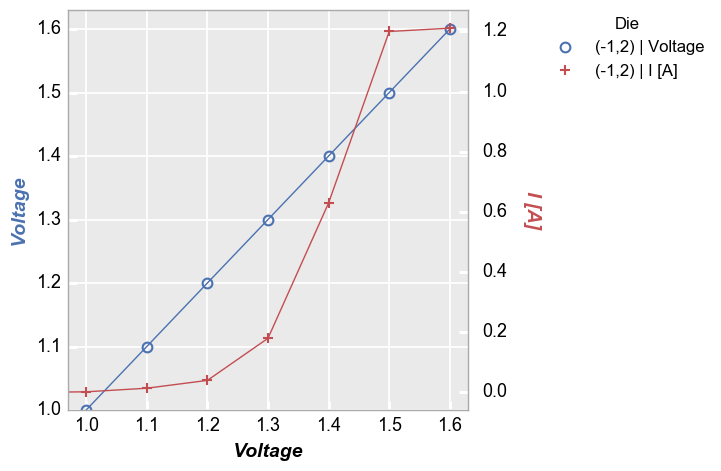
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"')
Now add limits to the shared x-axis:
In [10]:
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"', xmin=1.3)
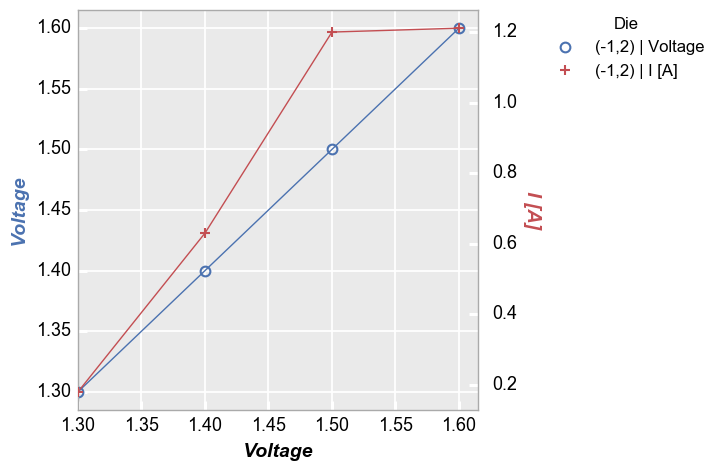
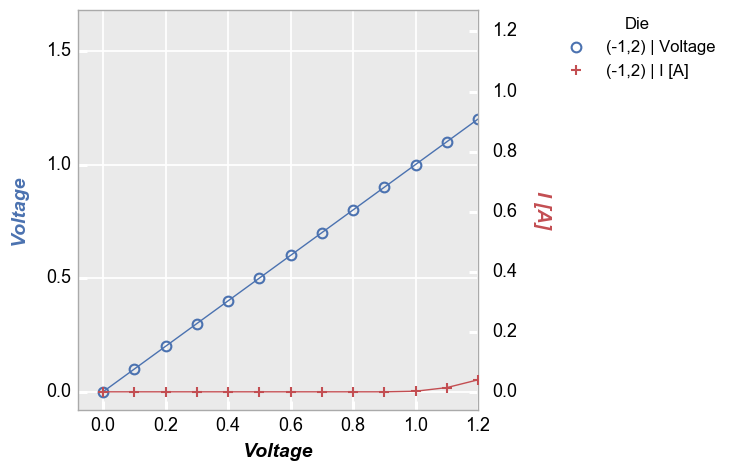
Because we have a shared x-axis in this case we see that both the primary and the seconday y-axis scale together when auto-scaling is enabled. As before we can disable auto-scaling if desired to treat the primary and secondary axes separately:
In [11]:
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"', xmax=1.2, auto_scale=False)
A similar auto-scaling effect will happen if we specify a y-limit (or a y2-limit). Again this is because the x-axis is shared and the auto-scaling algorithm filters the rows in the DataFrame based on our limits. Since both the primary
yand the secondaryyare columns in the same DataFrame, auto-scaling impacts both.In [12]:
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=True, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"', ymin=1)
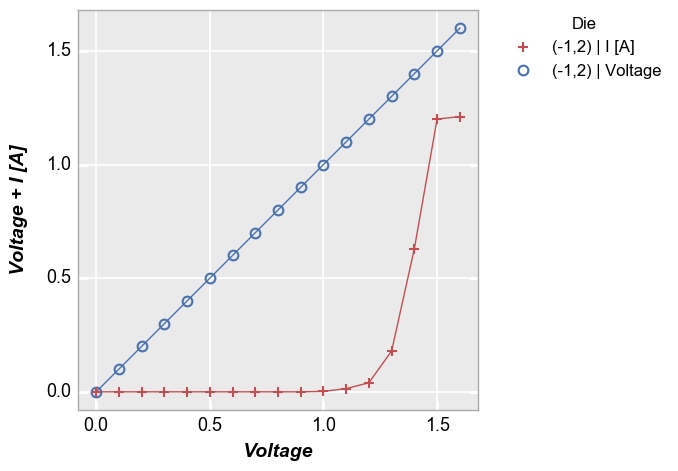
Multiple values on same axis¶
Lastly consider the non-twinned case with more than one value assigned to a given axis:
In [13]:
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=False, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"')
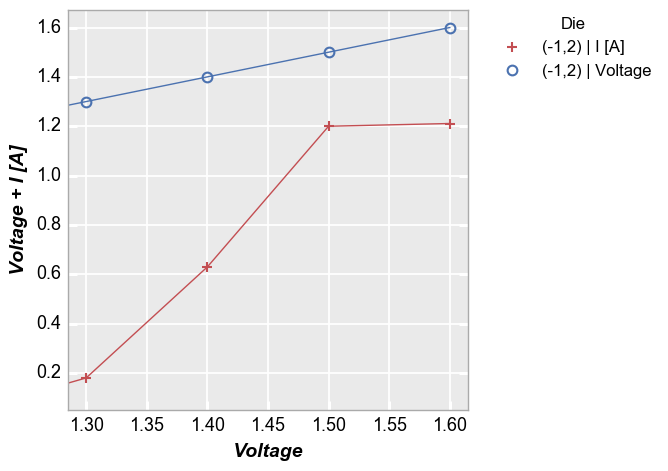
Here a y-limit is applied to both all the data on the y-axis so auto-scaling affects both curves:
In [14]:
fcp.plot(df=df, x='Voltage', y=['Voltage', 'I [A]'], twin_x=False, show=SHOW, legend='Die', filter='Substrate=="Si" & Target Wavelength==450 & Boost Level==0.2 & Temperature [C]==25 & Die=="(-1,2)"', ymin=0.05)
Note: auto-scaling is not active for boxplots, contours, and heatmaps.
Statistical limits¶
fivecentplots allows you to set axis limits based on some quantile percentage of the actual data or the inter-quartile range of the data. This is most useful when working with boxplots that contain outliers which we do not want to skew y-axis range.
Quantiles¶
Quantile ranges are added to the standard min/max keywords as strings
with the form: “<quantile>q”. Consider the plot below in which the
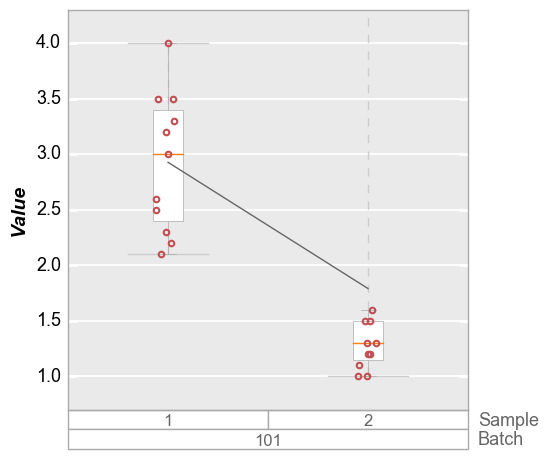
boxplot for sample 2 has an outlier. The default limit will cover the
entire span of the data so the ymax value is above this outlier.
In [15]:
fcp.boxplot(df=df_box, y='Value', groups=['Batch', 'Sample'], filter='Batch==101', show=SHOW)
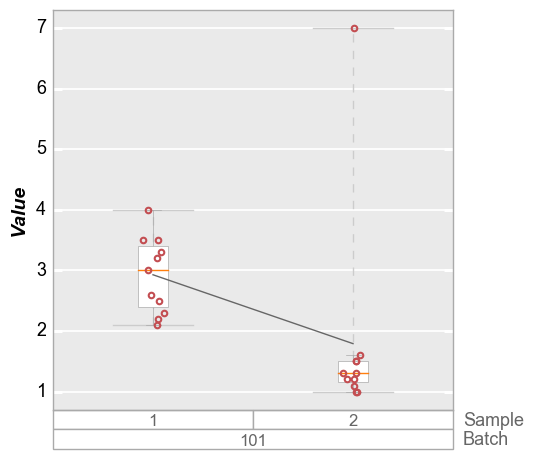
Obviously we could manually set a ymax value to exclude this
outlier, but in the case of automated plot generation we likely do not
know the outlier exists in advance. Instead we can specify a 95%
quantile limit to exclude tail points in the distribution. For boxplots,
if the range_lines option is enabled, we can still visualize that
there is an outlier in the data set that exceeds the y-axis range (see
here <boxplot.html#Range-lines>_)
In [16]:
fcp.boxplot(df=df_box, y='Value', groups=['Batch', 'Sample'], filter='Batch==101', show=SHOW, ymax='95q')
Inter-quartile range¶
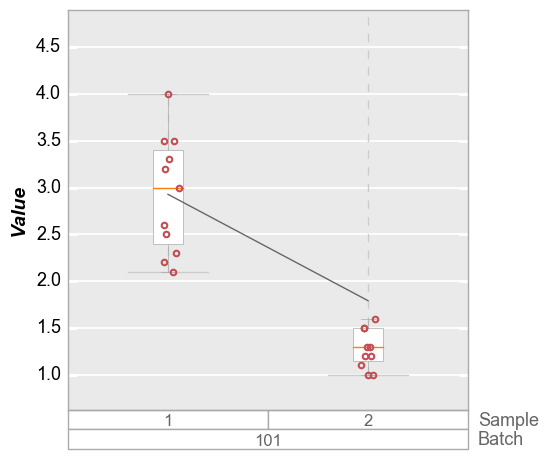
In some cases we may want to set a limit based on the inter-quartile range of the data set (i.e., the delta between the 25% and 75% quantiles). This can also help to deal with outlier data. The value supplied to the range keyword(s) is a string of the form: “<factor>*iqr”, where “factor” is a float value to be multiplied to the inter-quartile range.
In [17]:
fcp.boxplot(df=df_box, y='Value', groups=['Batch', 'Sample'], filter='Batch==101', show=SHOW,
ymin='1.5*iqr', ymax='1.5*iqr')
Axes sharing¶
Axes sharing applies when using row, col, or wrap grouping
to split the plot into multiple subplots. The boolean keywords of
interest are:
share_xshare_x2share_yshare_y2share_z