heatmap¶
This section provides examples of how to use the heatmap function. At a
minimum, the heatmap function requires the following keywords:
df: a pandas DataFramex: the name of the DataFrame column containing the x-axis datay: the name of the DataFrame column containing the y-axis dataz: the name of the DataFrame column containing the z-axis data
Heatmaps in fivecentplots can display both categorical and non-categorical data on either a uniform or non-uniform grid.
Setup¶
Imports¶
In [1]:
%load_ext autoreload
%autoreload 2
%matplotlib inline
import fivecentplots as fcp
import pandas as pd
import numpy as np
import os, sys, pdb
osjoin = os.path.join
db = pdb.set_trace
Sample data¶
In [2]:
df = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data_heatmap.csv'))
df.head()
Out[2]:
| Player | Category | Average | |
|---|---|---|---|
| 0 | Lebron James | Points | 27.5 |
| 1 | Lebron James | Assists | 9.1 |
| 2 | Lebron James | Rebounds | 8.6 |
| 3 | Lebron James | Blocks | 0.9 |
| 4 | James Harden | Points | 30.4 |
Set theme¶
In [3]:
#fcp.set_theme('gray')
#fcp.set_theme('white')
Other¶
In [4]:
SHOW = False
Categorical heatmap¶
First consider a case where both the x and y DataFrame columns
contain categorical data values:
No data labels¶
In [5]:
fcp.heatmap(df=df, x='Category', y='Player', z='Average', cbar=True, show=SHOW)
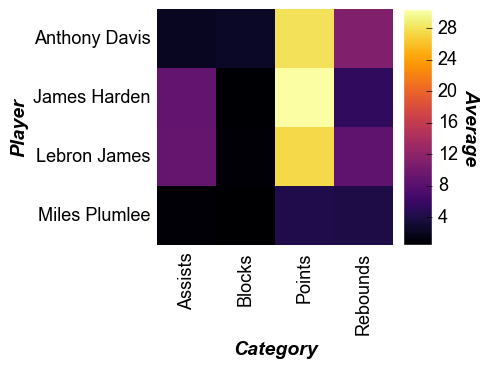
Note that for heatmaps the x tick labels are rotated 90° by default.
This can be overridden via the keyword tick_labels_major_x_rotation.
With data labels¶
In [6]:
fcp.heatmap(df=df, x='Category', y='Player', z='Average', cbar=True, data_labels=True,
heatmap_font_color='#aaaaaa', show=SHOW, tick_labels_major_y_edge_width=0, ws_ticks_ax=5)
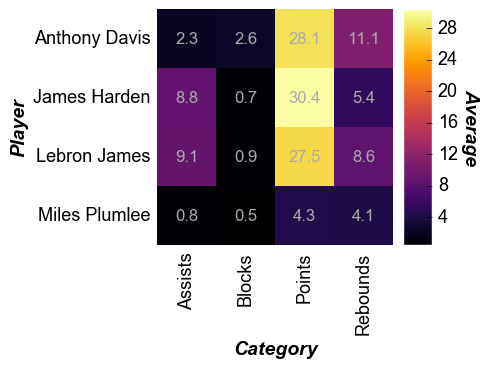
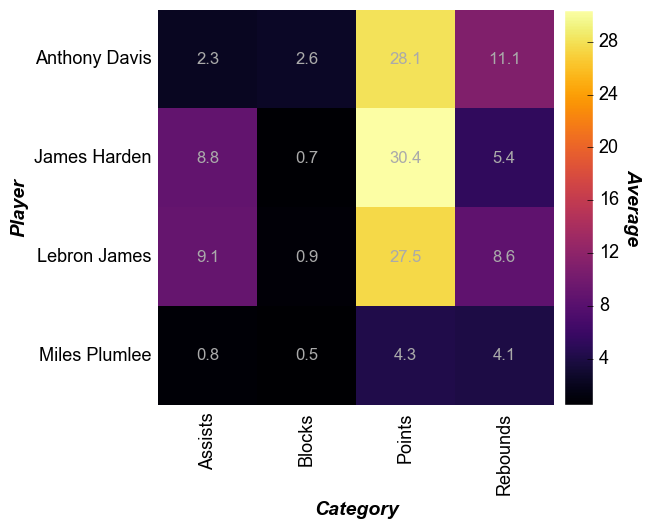
Cell size¶
The size of the heatmap cell will default to a width of 60 pixels
unless: (1) the keyword heatmap_cell_size (or cell_size when
directly supplying the value to the function call) is specified; or (2)
ax_size is explicitly defined. Note that for a heatmap the cells are
always square with width=height.
In [7]:
fcp.heatmap(df=df, x='Category', y='Player', z='Average', cbar=True, data_labels=True,
heatmap_font_color='#aaaaaa', show=SHOW, tick_labels_major_y_edge_width=0, ws_ticks_ax=5, cell_size=100)
Non-uniform data¶
A major difference between heatmaps and contour plots is that contour
plots assume that the x and y DataFrame column values are
numerical and continuous. With a heatmap, we can cast numerical data
into categorical form. Note that any missing values get mapped as
nan values are not not plotted.
In [8]:
# Read the contour DataFrame
df2 = pd.read_csv(osjoin(os.path.dirname(fcp.__file__), 'tests', 'fake_data_contour.csv'))
In [9]:
fcp.heatmap(df=df2, x='X', y='Y', z='Value', row='Batch', col='Experiment',
cbar=True, show=SHOW, share_z=True, ax_size=[400, 400],
data_labels=False, label_rc_font_size=12, filter='Batch==103', cmap='viridis')
Note that the x-axis width is not 400px as specified by the keyword
ax_scale. This occurs because the data set does not have as many
values on the x-axis as on the y-axis. fivecentplots applies the axis
size to the axis with the most items and scales the other axis
accordingly.
imshow alternative¶
We can also use fcp.heatmap to display images (similar to imshow
in matplotlib). Here we will take a random image from the world wide
web, place it in a pandas DataFrame, and display.
In [10]:
# Read an image
import imageio
url = 'https://s4827.pcdn.co/wp-content/uploads/2011/04/low-light-iphone4.jpg'
imgr = imageio.imread(url)
# Convert to grayscale
r, g, b = imgr[:,:,0], imgr[:,:,1], imgr[:,:,2]
gray = 0.2989 * r + 0.5870 * g + 0.1140 * b
# Convert image data to pandas DataFrame
img = pd.DataFrame(gray)
img.head()
Out[10]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 2582 | 2583 | 2584 | 2585 | 2586 | 2587 | 2588 | 2589 | 2590 | 2591 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.9999 | 0.0000 | 0.0000 | 2.2278 | 5.2275 | 5.2275 | 5.2275 | 4.2276 | 2.2278 | 2.2278 | ... | 0.8859 | 1.7718 | 2.7717 | 4.7715 | 4.7715 | 2.7717 | 2.0599 | 2.0599 | 2.0599 | 1.1740 |
| 1 | 0.9999 | 3.9996 | 2.9997 | 1.2279 | 5.2275 | 9.2271 | 7.2273 | 3.2277 | 2.2278 | 2.2278 | ... | 0.8859 | 0.8859 | 2.7717 | 1.7718 | 1.7718 | 3.7716 | 2.0599 | 0.5870 | 0.5870 | 1.1740 |
| 2 | 6.9993 | 4.9995 | 3.9996 | 2.2278 | 11.2269 | 18.2262 | 9.2271 | 0.2280 | 2.2278 | 1.2279 | ... | 6.7713 | 6.7713 | 2.7717 | 1.7718 | 2.7717 | 4.7715 | 4.0597 | 1.1740 | 0.5870 | 2.0599 |
| 3 | 13.9986 | 0.9999 | 1.9998 | 2.9997 | 7.2273 | 15.2265 | 9.2271 | 4.2276 | 1.2279 | 0.2280 | ... | 6.7713 | 6.7713 | 2.7717 | 4.7715 | 7.7712 | 8.7711 | 10.0591 | 9.0592 | 6.0595 | 3.0598 |
| 4 | 6.9993 | 0.0000 | 12.9987 | 14.9985 | 4.2276 | 5.2275 | 5.2275 | 4.2276 | 1.2279 | 0.2280 | ... | 1.7718 | 0.8859 | 3.7716 | 6.7713 | 10.7709 | 13.7706 | 16.0585 | 15.0586 | 10.0591 | 5.0596 |
5 rows × 2592 columns
Display the image as a colored heatmap:
In [11]:
fcp.heatmap(img, cmap='inferno', cbar=True, ax_size=[600, 600])

Now let’s enhance the contrast of the same image by limiting our color range to the mean pixel value +/- 3 * sigma:
In [12]:
uu = img.stack().mean()
ss = img.stack().std()
fcp.heatmap(img, cmap='inferno', cbar=True, ax_size=[600, 600], zmin=uu-3*ss, zmax=uu+3*ss)

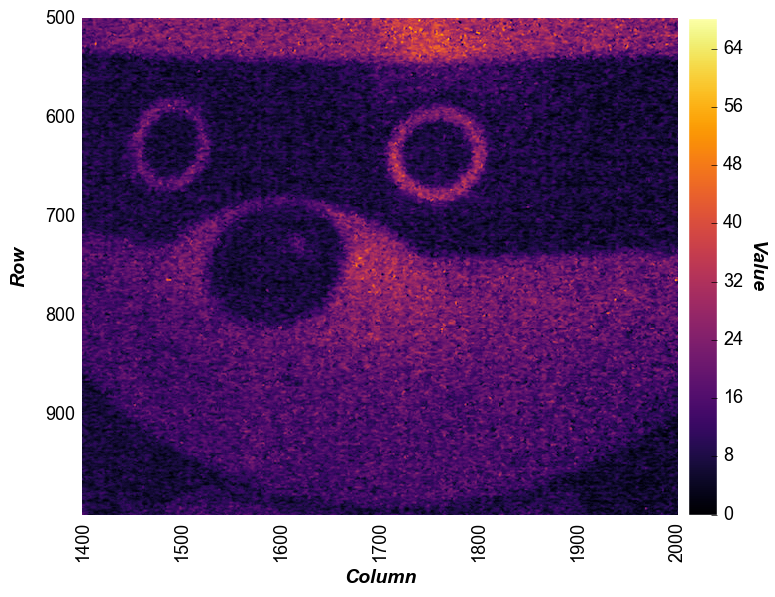
We can also crop the image by specifying range value for x and
y. Unlike imshow, the actual row and column values displayed on
the x- and y-axis are preserved after the zoom (not reset to 0, 0):
In [13]:
fcp.heatmap(img, cmap='inferno', cbar=True, ax_size=[600, 600], xmin=1400, xmax=2000, ymin=500, ymax=1000)
private eyes are watching you…